A1/ IMPORTING DATA
Choosing "Manage data" then "Import data" from the main horizontal menu leads to a page dedicated to data imports, split in two sections accessible via tabs named "Genotype import" and "Metadata import".
Anonymous users and users with no particular permissions are limited to importing genotyping data into temporary databases that remain accessible for 24h. These databases are created as public but hidden (only visible to people knowing their precise URL and to administrators).
From the second tab, users may thus import metadata (traits, phenotypes...) for a database's individuals. Metadata imported by users with write permissions (administrators, database supervisors, project managers) is considered "official" and therefore made available to all users by default. If specified by any other user, it will by only available for himself (thus completing / overriding the default metadata).
-
A1.1/ IMPORTING GENOTYPING DATA
Genotyping data may be provided in various formats (VCF, HapMap, PLINK, Intertek, FlapJack, DArTseq, BrAPI) and in various ways:
-
By specifying an absolute path on the webserver filesystem (convenient for administrators managing a production instance used as data portal);
-
By uploading files from the client computer (with an adjustable size limit: see section B8.2);
-
By providing an http URL, linking either to data files or to a BrAPI v1.1 base-url.
On the genotyping data import page, 3 more fields are required:
-
Database (not available to anonymous users since a "disposable" database is automatically generated for them): a database may contain one or several projects as long as they all rely on the same reference assembly. In the case of very large datasets, for performance-related matters it is however advised to have a single project per database;
-
Project: a project may contain one or several runs;
-
Run: each import process ends up adding a run to a project. Allowing multiple runs in a project is a way of supporting incremental data loading.
For each individual referred to from an import file (e.g., VCF), a sample is added to the database. Thus, this allows accounting for multiple samples for a same individual. By default, samples are given an auto-generated name, but it is possible to make Gigwa consider the names featured in import files as sample IDs rather than individual IDs (useful when using BrAPI for browsing data). This is achieved by checking the box labelled "Genotypes provided for samples, not individuals". Three different cases are then supported for naming individuals: (1) if no additional info is provided, individuals will be named same as samples; (2) from the "Genotype import" tab, users may supply an additional 2-column tabulated file listing ID mappings (with a header featuring 'individual' and 'sample'); (3) via BrAPI from the "Metadata import" tab" (explained below at A1.3).
Specifying a ploidy level is optional (most of the time, the system is able to guess it) but recommended for Flapjack and HapMap formats (will speed-up imports). A descriptive text can be provided for each project. A tooltipped lightbulb icon explains how to add a how-to-cite text that would then be exported along with any data extracted from the given project.

Genotyping data import progress can be watched in real time from the upload page, or via a dedicated asynchronous progress page (convenient for large datasets), while imports run as background processes.
-
-
A1.2/ IMPORTING METADATA, the classical way
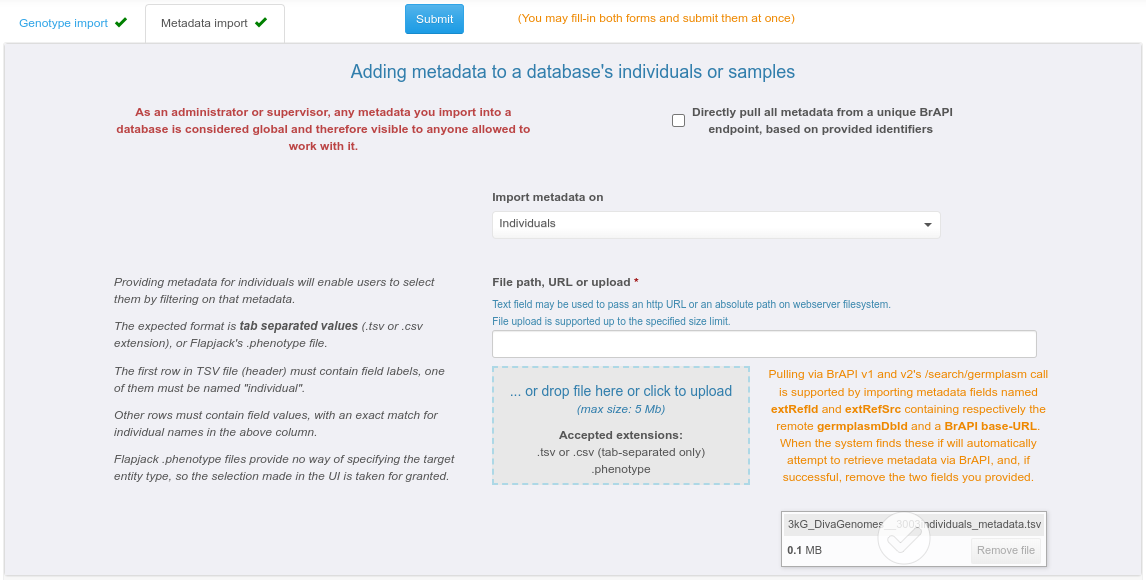
Providing metadata may be done either for existing individuals / samples, or while importing them along with genotypes. Individual-level metadata aims at enabling users to batch-select them by filtering on it. This is convenient for cases where the individual list is long and / or individual names are not meaningful. Sample-level metadata can also be stored by the system but is currently not reflected in the default Gigwa UI (only accessible via BrAPI).
Metadata may be provided as a simple tabulated file containing user-defined columns (only one is enforced, named 'individual' or 'sample' depending on the selected target entities).
Importing metadata via BrAPI v1 or v2 is also supported. To do that, you must submit a tabulated metadata file with at least the 2 following columns (in addition to above-mentioned mandatory column):
-
extRefSrc which must contain the BrAPI url (e.g. https://test-server.brapi.org/brapi/v2) to pull entity metadata from;
-
extRefId which must contain the germplasmDbId or sampleDbId (on the BrAPI server) corresponding to the Gigwa individual or sample respectively referred to on the current line.
As a different extRefSrc may be provided on each line, it is possible to pull data from multiple BrAPI sources at once (user will be prompted for an optional token to use with each of them). In the end, Gigwa will attempt to grab germplasm or sample metadata from BrAPI to supplement user-provided information upon form submission.
-
-
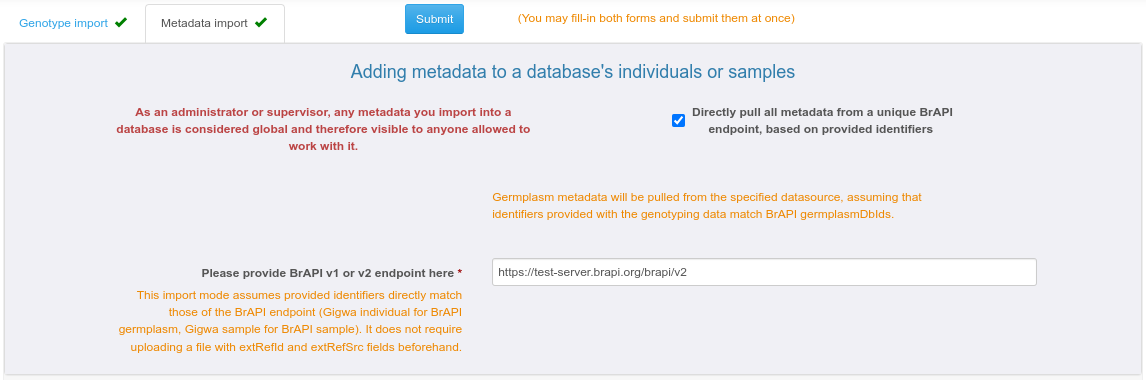
A1.3/ IMPORTING METADATA at once from BrAPI, when provided identifiers match those in the BrAPI source
Providing metadata based on matching identifiers can be achieved by checking the "Directly pull all metadata from a unique BrAPI endpoint, based on provided identifier" box.
In this case, assuming that individuals in Gigwa correspond to germplasm in BrAPI and that names provided for biological material through the genotyping data file(s) match BrAPI dbIds, Gigwa just needs a valid BrAPI endpoint to populate metadata from its contents.
If the "Genotypes provided for samples, not individuals" box is checked in the first tab, the system will query the BrAPI endpoint to find all necessary germplasm information, and thus name individuals based on this.